EasySep™ Human Myeloid DC Enrichment Kit
Immunomagnetic negative selection cell isolation kit
概要
The EasySep™ Human Myeloid DC Enrichment Kit is designed to isolate myeloid dendritic cells (mDC) from fresh peripheral blood mononuclear cells or ammonium chloride-lysed leukapheresis by negative selection. Unwanted cells are targeted for removal with Tetrameric Antibody Complexes recognizing non-mDCs and dextran-coated magnetic particles. The labeled cells are separated using the EasySep™ magnet without the use of columns. Desired cells are poured off into a new tube.
Advantages
• Fast, easy-to-use and column-free
• Up to 90% purity
• Untouched, viable cells
• Up to 90% purity
• Untouched, viable cells
Components
- EasySep™ Human Myeloid DC Enrichment Kit (Catalog #19061)
- EasySep™ Human Myeloid DC Enrichment Cocktail Component A, 2 x 1 mL
- EasySep™ Human DC Enrichment Cocktail Component B, 2 x 1 mL
- EasySep™ D Magnetic Particles, 5 x 1 mL
- Anti-Human CD32 (Fc gamma RII) Blocker, 0.8 mL
- RoboSep™ Human Myeloid DC Enrichment Kit (Catalog #19061RF)
- EasySep™ Human Myeloid DC Enrichment Cocktail Component A, 2 x 1 mL
- EasySep™ Human DC Enrichment Cocktail Component B, 2 x 1 mL
- EasySep™ D Magnetic Particles, 5 x 1 mL
- Anti-Human CD32 (Fc gamma RII) Blocker, 0.8 mL
- RoboSep™ Buffer (Catalog #20104) x 2
- RoboSep™ Filter Tips (Catalog #20125) x 2
Magnet Compatibility
• EasySep™ Magnet (Catalog #18000)
• “The Big Easy” EasySep™ Magnet (Catalog #18001)
• RoboSep™-S (Catalog #21000)
Subtype
Cell Isolation Kits
Cell Type
Dendritic Cells
Species
Human
Sample Source
PBMC
Selection Method
Negative
Application
Cell Isolation
Brand
EasySep, RoboSep
Area of Interest
Immunology
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | EasySep™ Human Myeloid DC Enrichment Kit | 19061 | All | English |
| Product Information Sheet | RoboSep™ Human Myeloid DC Enrichment Kit | 19061RF | All | English |
| Safety Data Sheet 1 | EasySep™ Human Myeloid DC Enrichment Kit | 19061 | All | English |
| Safety Data Sheet 2 | EasySep™ Human Myeloid DC Enrichment Kit | 19061 | All | English |
| Safety Data Sheet 1 | RoboSep™ Human Myeloid DC Enrichment Kit | 19061RF | All | English |
| Safety Data Sheet 2 | RoboSep™ Human Myeloid DC Enrichment Kit | 19061RF | All | English |
数据及文献
Data
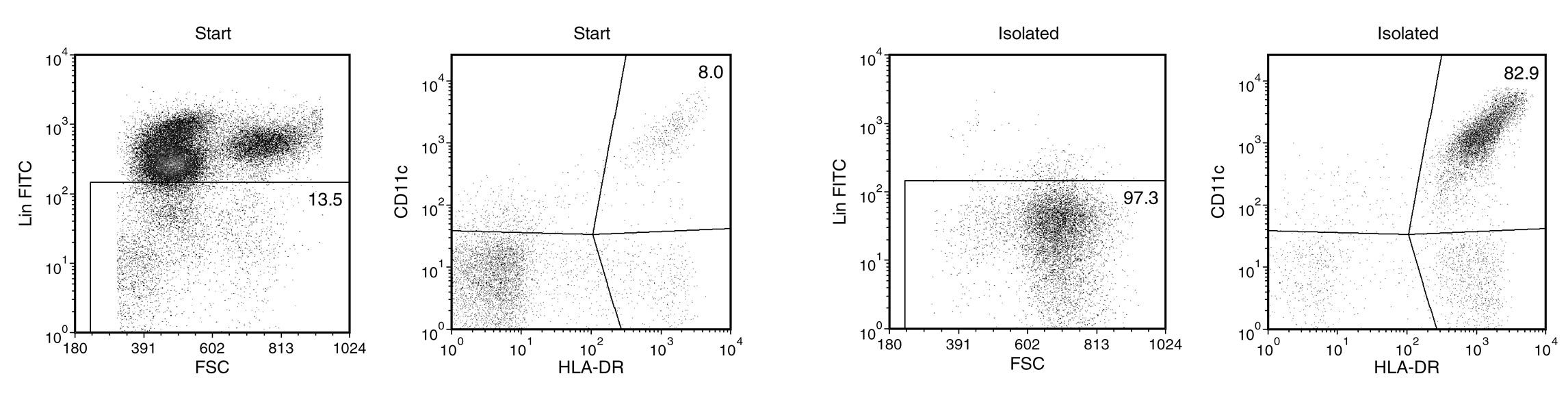
Figure 1. EasySep™ Human Myeloid DC Enrichment Kit
Starting with 0.6 - 1.8% mDCs in fresh peripheral blood nucleated cells, the mDC content of the enriched fraction typically ranges from 50 - 90%* purity based on the mDC phenotype of Lineage (CD3, CD14, CD19, CD20, CD34, CD56)-negative, HLA DR-positive, and CD11c-positive. In the above example, the purities of the start and final enriched fractions are 1.1% and 80.7%, respectively. *If the mDC content of the starting sample is < 1.25%, the mDC content of the enriched fraction may be < 80%.
Publications (2)
Blood 2017
NOX5 and p22phox are 2 novel regulators of human monocytic differentiation into dendritic cells.
Abstract
Abstract
Dendritic cells (DCs) are a heterogeneous population of professional antigen-presenting cells and are key cells of the immune system, acquiring different phenotypes in accordance with their localization during the immune response. A subset of inflammatory DCs is derived from circulating monocytes (Mo) and has a key role in inflammation and infection. The pathways controlling Mo-DC differentiation are not fully understood. Our objective was to investigate the possible role of nicotinamide adenine dinucleotide phosphate reduced form oxidases (NOXs) in Mo-DC differentiation. In this study, we revealed that Mo-DC differentiation was inhibited by NOX inhibitors and reactive oxygen species scavengers. We show that the Mo-DC differentiation was dependent on p22phox, and not on gp91phox/NOX2, as shown by the reduced Mo-DC differentiation observed in chronic granulomatous disease patients lacking p22phox. Moreover, we revealed that NOX5 expression was strongly increased during Mo-DC differentiation, but not during Mo-macrophage differentiation. NOX5 was expressed in circulating myeloid DC, and at a lower level in plasmacytoid DC. Interestingly, NOX5 was localized at the outer membrane of the mitochondria and interacted with p22phox in Mo-DC. Selective inhibitors and small interfering RNAs for NOX5 indicated that NOX5 controlled Mo-DC differentiation by regulating the JAK/STAT/MAPK and NFκB pathways. These data demonstrate that the NOX5-p22phox complex drives Mo-DC differentiation, and thus could be critical for immunity and inflammation.
PloS one 2012
Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression.
Abstract
Abstract
Blood consists of different cell populations with distinct functions and correspondingly, distinct gene expression profiles. In this study, global miRNA expression profiling was performed across a panel of nine human immune cell subsets (neutrophils, eosinophils, monocytes, B cells, NK cells, CD4 T cells, CD8 T cells, mDCs and pDCs) to identify cell-type specific miRNAs. mRNA expression profiling was performed on the same samples to determine if miRNAs specific to certain cell types down-regulated expression levels of their target genes. Six cell-type specific miRNAs (miR-143; neutrophil specific, miR-125; T cells and neutrophil specific, miR-500; monocyte and pDC specific, miR-150; lymphoid cell specific, miR-652 and miR-223; both myeloid cell specific) were negatively correlated with expression of their predicted target genes. These results were further validated using an independent cohort where similar immune cell subsets were isolated and profiled for both miRNA and mRNA expression. miRNAs which negatively correlated with target gene expression in both cohorts were identified as candidates for miRNA/mRNA regulatory pairs and were used to construct a cell-type specific regulatory network. miRNA/mRNA pairs formed two distinct clusters in the network corresponding to myeloid (nine miRNAs) and lymphoid lineages (two miRNAs). Several myeloid specific miRNAs targeted common genes including ABL2, EIF4A2, EPC1 and INO80D; these common targets were enriched for genes involved in the regulation of gene expression (ptextless9.0E-7). Those miRNA might therefore have significant further effect on gene expression by repressing the expression of genes involved in transcriptional regulation. The miRNA and mRNA expression profiles reported in this study form a comprehensive transcriptome database of various human blood cells and serve as a valuable resource for elucidating the role of miRNA mediated regulation in the establishment of immune cell identity.