EasySep™ Human Memory CD4+ T Cell Enrichment Kit
Immunomagnetic negative selection kit
概要
The EasySep™ Human Memory CD4 T Cell Enrichment Kit is designed to isolate memory CD4+ T cells from fresh or previously frozen peripheral blood mononuclear cells by negative selection. Unwanted cells are targeted for removal with Tetrameric Antibody Complexes recognizing CD8, CD14, CD16, CD19, CD20, CD36, CD45RA, CD56, CD123, TCRγ/δ, glycophorin A and dextran-coated magnetic particles. The labeled cells are separated using an EasySep™ magnet without the use of columns. Desired cells are poured off into a new tube.
Advantages
• Fast, easy-to-use and column-free
• Up to 98% purity
• Untouched, viable cells
• Up to 98% purity
• Untouched, viable cells
Components
- EasySep™ Human Memory CD4+ T Cell Enrichment Kit (Catalog #19157)
- EasySep™ Human Memory CD4+ T Cell Enrichment Cocktail, 1 mL
- EasySep™ D Magnetic Particles, 2 x 1 mL
- RoboSep™ Human Memory CD4 T Cell Enrichment Kit with Filter Tips (Catalog #19157RF)
- EasySep™ Human Memory CD4+ T Cell Enrichment Cocktail, 1 mL
- EasySep™ D Magnetic Particles, 2 x 1 mL
- RoboSep™ Buffer (Catalog #20104)
- RoboSep™ Filter Tips (Catalog #20125)
Magnet Compatibility
• EasySep™ Magnet (Catalog #18000)
• “The Big Easy” EasySep™ Magnet (Catalog #18001)
• EasyPlate™ EasySep™ Magnet (Catalog 18102)
• Easy 50 EasySep™ Magnet (Catalog #18002)
• RoboSep™-S (Catalog #21000)
Subtype
Cell Isolation Kits
Cell Type
T Cells, T Cells, CD4+
Species
Human
Sample Source
PBMC
Selection Method
Negative
Application
Cell Isolation
Brand
EasySep, RoboSep
Area of Interest
Immunology
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | EasySep™ Human Memory CD4+ T Cell Enrichment Kit | 19157 | All | English |
| Product Information Sheet | RoboSep™ Human Memory CD4 T Cell Enrichment Kit with Filter Tips | 19157RF | All | English |
| Safety Data Sheet 1 | EasySep™ Human Memory CD4+ T Cell Enrichment Kit | 19157 | All | English |
| Safety Data Sheet 2 | EasySep™ Human Memory CD4+ T Cell Enrichment Kit | 19157 | All | English |
| Safety Data Sheet 3 | EasySep™ Human Memory CD4+ T Cell Enrichment Kit | 19157 | All | English |
| Safety Data Sheet 1 | RoboSep™ Human Memory CD4 T Cell Enrichment Kit with Filter Tips | 19157RF | All | English |
| Safety Data Sheet 2 | RoboSep™ Human Memory CD4 T Cell Enrichment Kit with Filter Tips | 19157RF | All | English |
| Safety Data Sheet 3 | RoboSep™ Human Memory CD4 T Cell Enrichment Kit with Filter Tips | 19157RF | All | English |
数据及文献
Data
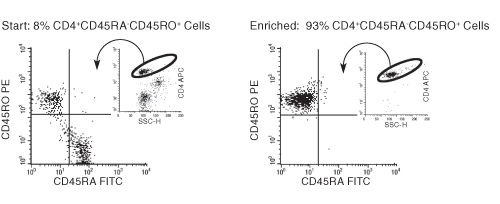
Figure 1. Typical Enrichment Profile For EasySep™ Human Memory CD4+ T Cell Enrichment Kit
Starting with previously frozen mononuclear cells, the memory CD4+ T cell content of the enriched fraction typically ranges from 86% - 98%.
Publications (3)
Nature communications 2019 feb
PD-1 blockade potentiates HIV latency reversal ex vivo in CD4+ T cells from ART-suppressed individuals.
Abstract
Abstract
HIV persists in latently infected CD4+ T cells during antiretroviral therapy (ART). Immune checkpoint molecules, including PD-1, are preferentially expressed at the surface of persistently infected cells. However, whether PD-1 plays a functional role in HIV latency and reservoir persistence remains unknown. Using CD4+ T cells from HIV-infected individuals, we show that the engagement of PD-1 inhibits viral production at the transcriptional level and abrogates T-cell receptor (TCR)-induced HIV reactivation in latently infected cells. Conversely, PD-1 blockade with the monoclonal antibody pembrolizumab enhances HIV production in combination with the latency reversing agent bryostatin without increasing T cell activation. Our results suggest that the administration of immune checkpoint blockers to HIV-infected individuals on ART may facilitate latency disruption.
Nature communications 2019
High-throughput single-cell rheology in complex samples by dynamic real-time deformability cytometry.
Abstract
Abstract
In life sciences, the material properties of suspended cells have attained significance close to that of fluorescent markers but with the advantage of label-free and unbiased sample characterization. Until recently, cell rheological measurements were either limited by acquisition throughput, excessive post processing, or low-throughput real-time analysis. Real-time deformability cytometry expanded the application of mechanical cell assays to fast on-the-fly phenotyping of large sample sizes, but has been restricted to single material parameters as the Young's modulus. Here, we introduce dynamic real-time deformability cytometry for comprehensive cell rheological measurements at up to 100 cells per second. Utilizing Fourier decomposition, our microfluidic method is able to disentangle cell response to complex hydrodynamic stress distributions and to determine viscoelastic parameters independent of cell shape. We demonstrate the application of our technology for peripheral blood cells in whole blood samples including the discrimination of B- and CD4+ T-lymphocytes by cell rheological properties.
Nature Communications 2015 DEC
Intermediate DNA methylation is a conserved signature of genome regulation
Abstract
Abstract
The role of intermediate methylation states in DNA is unclear. Here, to comprehensively identify regions of intermediate methylation and their quantitative relationship with gene activity, we apply integrative and comparative epigenomics to 25 human primary cell and tissue samples. We report 18,452 intermediate methylation regions located near 36% of genes and enriched at enhancers, exons and DNase I hypersensitivity sites. Intermediate methylation regions average 57% methylation, are predominantly allele-independent and are conserved across individuals and between mouse and human, suggesting a conserved function. These regions have an intermediate level of active chromatin marks and their associated genes have intermediate transcriptional activity. Exonic intermediate methylation correlates with exon inclusion at a level between that of fully methylated and unmethylated exons, highlighting gene context-dependent functions. We conclude that intermediate DNA methylation is a conserved signature of gene regulation and exon usage.