EasySep™ Human CD4+ T Cell Isolation Kit
8-Minute cell isolation kit using immunomagnetic negative selection
概要
The EasySep™ Human CD4+ T Cell Isolation Kit is designed to isolate CD4+ T cells from fresh or previously frozen peripheral blood mononuclear cells or washed leukapheresis samples by immunomagnetic negative selection. The EasySep™ procedure involves labeling unwanted cells with antibody complexes and magnetic particles. The magnetically labeled cells are separated from the untouched desired cells by using an EasySep™ magnet and simply pouring or pipetting the desired cells into a new tube.
This product can be used in place of the EasySep™ Human CD4+ T Cell Enrichment Kit (Catalog #19052) for even faster cell isolations.
This product can be used in place of the EasySep™ Human CD4+ T Cell Enrichment Kit (Catalog #19052) for even faster cell isolations.
Advantages
• Fast, easy-to-use and column-free
• Up to 97% purity with high recovery
• Untouched, viable cells
• Up to 97% purity with high recovery
• Untouched, viable cells
Components
- EasySep™ Human CD4+ T Cell Isolation Kit (Catalog #17952)
- EasySep™ Human CD4+ T Cell Isolation Cocktail, 1 mL
- EasySep™ Dextran RapidSpheres™, 1 mL
- RoboSep™ Human CD4+ T Cell Isolation Kit (Catalog #17952RF)
- EasySep™ Human CD4+ T Cell Isolation Cocktail, 1 mL
- EasySep™ Dextran RapidSpheres™, 1 mL
- RoboSep™ Buffer (Catalog #20104)
- RoboSep™ Filter Tips (Catalog #20125)
Magnet Compatibility
• EasySep™ Magnet (Catalog #18000)
• “The Big Easy” EasySep™ Magnet (Catalog #18001)
• Easy 50 EasySep™ Magnet (Catalog #18002)
• EasyPlate™ EasySep™ Magnet (Catalog 18102)
• EasyEights™ EasySep™ Magnet (Catalog #18103)
• RoboSep™-S (Catalog #21000)
Subtype
Cell Isolation Kits
Cell Type
T Cells, T Cells, CD4+
Species
Human
Sample Source
Leukapheresis, PBMC
Selection Method
Negative
Application
Cell Isolation
Brand
EasySep, RoboSep
Area of Interest
Immunology
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | EasySep™ Human CD4+ T Cell Isolation Kit | 17952 | All | English |
| Product Information Sheet | RoboSep™ Human CD4+ T Cell Isolation Kit | 17952RF | All | English |
| Safety Data Sheet 1 | EasySep™ Human CD4+ T Cell Isolation Kit | 17952 | All | English |
| Safety Data Sheet 2 | EasySep™ Human CD4+ T Cell Isolation Kit | 17952 | All | English |
| Safety Data Sheet 1 | RoboSep™ Human CD4+ T Cell Isolation Kit | 17952RF | All | English |
| Safety Data Sheet 2 | RoboSep™ Human CD4+ T Cell Isolation Kit | 17952RF | All | English |
数据及文献
Data
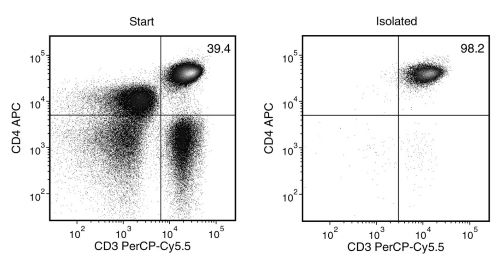
Figure 1. EasySep™ Human CD4+ T Cell Isolation Kit
Starting with human peripheral blood mononuclear cells (PBMCs), the CD4+ T cell (CD3+CD4+) content of the isolated fraction is typically 94.8 ± 2.3% (mean ± SD).
Publications (23)
Science advances 2020 may
Competition between PAF1 and MLL1/COMPASS confers the opposing function of LEDGF/p75 in HIV latency and proviral reactivation.
Abstract
Abstract
Transcriptional status determines the HIV replicative state in infected patients. However, the transcriptional mechanisms for proviral replication control remain unclear. In this study, we show that, apart from its function in HIV integration, LEDGF/p75 differentially regulates HIV transcription in latency and proviral reactivation. During latency, LEDGF/p75 suppresses proviral transcription via promoter-proximal pausing of RNA polymerase II (Pol II) by recruiting PAF1 complex to the provirus. Following latency reversal, MLL1 complex competitively displaces PAF1 from the provirus through casein kinase II (CKII)-dependent association with LEDGF/p75. Depleting or pharmacologically inhibiting CKII prevents PAF1 dissociation and abrogates the recruitment of both MLL1 and Super Elongation Complex (SEC) to the provirus, thereby impairing transcriptional reactivation for latency reversal. These findings, therefore, provide a mechanistic understanding of how LEDGF/p75 coordinates its distinct regulatory functions at different stages of the post-integrated HIV life cycles. Targeting these mechanisms may have a therapeutic potential to eradicate HIV infection.
Cancer research 2019 may
Inhibition of EphB4-Ephrin-B2 Signaling Reprograms the Tumor Immune Microenvironment in Head and Neck Cancers.
Abstract
Abstract
Identifying targets present in the tumor microenvironment that contribute to immune evasion has become an important area of research. In this study, we identified EphB4-ephrin-B2 signaling as a regulator of both innate and adaptive components of the immune system. EphB4 belongs to receptor tyrosine kinase family that interacts with ephrin-B2 ligand at sites of cell-cell contact, resulting in bidirectional signaling. We found that EphB4-ephrin-B2 inhibition alone or in combination with radiation (RT) reduced intratumoral regulatory T cells (Tregs) and increased activation of both CD8+ and CD4+Foxp3- T cells compared with the control group in an orthotopic head and neck squamous cell carcinoma (HNSCC) model. We also compared the effect of EphB4-ephrin-B2 inhibition combined with RT with combined anti-PDL1 and RT and observed similar tumor growth suppression, particularly at early time-points. A patient-derived xenograft model showed reduction of tumor-associated M2 macrophages and favored polarization towards an antitumoral M1 phenotype following EphB4-ephrin-B2 inhibition with RT. In vitro, EphB4 signaling inhibition decreased Ki67-expressing Tregs and Treg activation compared with the control group. Overall, our study is the first to implicate the role of EphB4-ephrin-B2 in tumor immune response. Moreover, our findings suggest that EphB4-ephrin-B2 inhibition combined with RT represents a potential alternative for patients with HNSCC and could be particularly beneficial for patients who are ineligible to receive or cannot tolerate anti-PDL1 therapy. SIGNIFICANCE: These findings present EphB4-ephrin-B2 inhibition as an alternative to anti-PDL1 therapeutics that can be used in combination with radiation to induce an effective antitumor immune response in patients with HNSCC.
Science advances 2019 mar
Hybrid nanocarriers incorporating mechanistically distinct drugs for lymphatic CD4+ T cell activation and HIV-1 latency reversal.
Abstract
Abstract
A proposed strategy to cure HIV uses latency-reversing agents (LRAs) to reactivate latent proviruses for purging HIV reservoirs. A variety of LRAs have been identified, but none has yet proven effective in reducing the reservoir size in vivo. Nanocarriers could address some major challenges by improving drug solubility and safety, providing sustained drug release, and simultaneously delivering multiple drugs to target tissues and cells. Here, we formulated hybrid nanocarriers that incorporate physicochemically diverse LRAs and target lymphatic CD4+ T cells. We identified one LRA combination that displayed synergistic latency reversal and low cytotoxicity in a cell model of HIV and in CD4+ T cells from virologically suppressed patients. Furthermore, our targeted nanocarriers selectively activated CD4+ T cells in nonhuman primate peripheral blood mononuclear cells as well as in murine lymph nodes, and substantially reduced local toxicity. This nanocarrier platform may enable new solutions for delivering anti-HIV agents for an HIV cure.
Nature microbiology 2019 feb
HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy.
Abstract
Abstract
Human immunodeficiency virus type 1 (HIV-1) eradication is prevented by the establishment on infection of cellular HIV-1 reservoirs that are not fully characterized, especially in genital mucosal tissues (the main HIV-1 entry portal on sexual transmission). Here, we show, using penile tissues from HIV-1-infected individuals under suppressive combination antiretroviral therapy, that urethral macrophages contain integrated HIV-1 DNA, RNA, proteins and intact virions in virus-containing compartment-like structures, whereas viral components remain undetectable in urethral T cells. Moreover, urethral cells specifically release replication-competent infectious HIV-1 following reactivation with the macrophage activator lipopolysaccharide, while the T-cell activator phytohaemagglutinin is ineffective. HIV-1 urethral reservoirs localize preferentially in a subset of polarized macrophages that highly expresses the interleukin-1 receptor, CD206 and interleukin-4 receptor, but not CD163. To our knowledge, these results are the first evidence that human urethral tissue macrophages constitute a principal HIV-1 reservoir. Such findings are determinant for therapeutic strategies aimed at HIV-1 eradication.
The Journal of clinical investigation 2019 dec
Myalgic encephalomyelitis/chronic fatigue syndrome patients exhibit altered T cell metabolism and cytokine associations.
Abstract
Abstract
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a complex disease with no known cause or mechanism. There is an increasing appreciation for the role of immune and metabolic dysfunction in the disease. ME/CFS has historically presented in outbreaks, often has a flu-like onset, and results in inflammatory symptoms. Patients suffer from severe fatigue and post-exertional malaise. There is little known about the metabolism of specific immune cells in ME/CFS patients. To investigate immune metabolism in ME/CFS, we isolated CD4+ and CD8+ T cells from 53 ME/CFS patients and 45 healthy controls. We analyzed glycolysis and mitochondrial respiration in resting and activated T cells, along with markers related to cellular metabolism, and plasma cytokines. We found that ME/CFS CD8+ T cells have reduced mitochondrial membrane potential compared to healthy controls. Both CD4+ and CD8+ T cells from ME/CFS patients had reduced glycolysis at rest, while CD8+ T cells also had reduced glycolysis following activation. ME/CFS patients had significant correlations between measures of T cell metabolism and plasma cytokine abundance that differed from healthy control subjects. Our data indicate that patients have impaired T cell metabolism consistent with ongoing immune alterations in ME/CFS that may illuminate the mechanism behind this disease.
Cell and tissue research 2019 dec
Characterizing the effects of hypoxia on the metabolic profiles of mesenchymal stromal cells derived from three tissue sources using chemical isotope labeling liquid chromatography-mass spectrometry.
Abstract
Abstract
Microenvironmental factors such as oxygen concentration mediate key effects on the biology of mesenchymal stromal cells (MSCs). Herein, we performed an in-depth characterization of the metabolic behavior of MSCs derived from the placenta, umbilical cord, and adipose tissue (termed hPMSCs, UC-MSCs, and AD-MSCs, respectively) at physiological (hypoxic; 5{\%} oxygen [O2]) and standardized (normoxic; 21{\%} O2) O2 concentrations using chemical isotope labeling liquid chromatography-mass spectrometry. 12C- and 13C-isotope dansylation (Dns) labeling was used to analyze the amine/phenol submetabolome, and 2574 peak pairs or metabolites were detected and quantified, from which 52 metabolites were positively identified using a library of 275 Dns-metabolite standards; 2189 metabolites were putatively identified. Next, we identified six metabolites using the Dns library, as well as 14 hypoxic biomarkers from the human metabolome database out of 96 altered metabolites. Ultimately, metabolic pathway analyses were performed to evaluate the associated pathways. Based on pathways identified using the Kyoto Encyclopedia of Genes and Genomes, we identified significant changes in the metabolic profiles of MSCs in response to different O2 concentrations. These results collectively suggest that O2 concentration has the strongest influence on hPMSCs metabolic characteristics, and that 5{\%} O2 promotes arginine and proline metabolism in hPMSCs and UC-MSCs but decreases gluconeogenesis (alanine-glucose) rates in hPMSCs and AD-MSCs. These changes indicate that MSCs derived from different sources exhibit distinct metabolic profiles.