EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit
Immunomagnetic negative selection cell isolation kit
概要
The EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit is designed for the depletion of single or multiple unwanted cell types labeled with biotinylated antibodies. Labeled cells from mouse splenocytes or other tissues are targeted for removal by Streptavidin RapidSpheres™ and separated without the use of columns using an EasySep™ magnet. Desired cells are untouched and poured off into a new tube.
Advantages
• Fast and easy-to-use
• No columns required
• Untouched, viable cells
• No columns required
• Untouched, viable cells
Components
- EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit (Catalog #19860)
- EasySep™ Streptavidin RapidSpheres™ 50001, 1 mL
- Normal Rat Serum, 2 mL
- RoboSep™ Empty Vial
- RoboSep™ Mouse Streptavidin RapidSpheres™ Isolation Kit (Catalog #19860RF)
- EasySep™ Streptavidin RapidSpheres™ 50001, 1 mL
- Normal Rat Serum, 2 mL
- RoboSep™ Empty Vial
- RoboSep™ Buffer (Catalog #20104)
- RoboSep™ Filter Tips (Catalog #20125)
Magnet Compatibility
• EasySep™ Magnet (Catalog #18000)
• “The Big Easy” EasySep™ Magnet (Catalog #18001)
• RoboSep™-S (Catalog #21000)
Subtype
Cell Isolation Kits
Cell Type
B Cells, Dendritic Cells, Granulocytes and Subsets, Hematopoietic Stem and Progenitor Cells, Macrophages, Marrow Stromal Cells, Mesenchymal Stem and Progenitor Cells, Monocytes, Mononuclear Cells, Myeloid Cells, NK Cells, Other, Plasma, T Cells
Species
Mouse
Sample Source
Other, Spleen
Selection Method
Depletion, Negative
Application
Cell Isolation
Brand
EasySep, RoboSep
Area of Interest
Immunology
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860 | All | English |
| Product Information Sheet | RoboSep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860RF | All | English |
| Safety Data Sheet 1 | EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860 | All | English |
| Safety Data Sheet 2 | EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860 | All | English |
| Safety Data Sheet 1 | RoboSep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860RF | All | English |
| Safety Data Sheet 2 | RoboSep™ Mouse Streptavidin RapidSpheres™ Isolation Kit | 19860RF | All | English |
数据及文献
Data
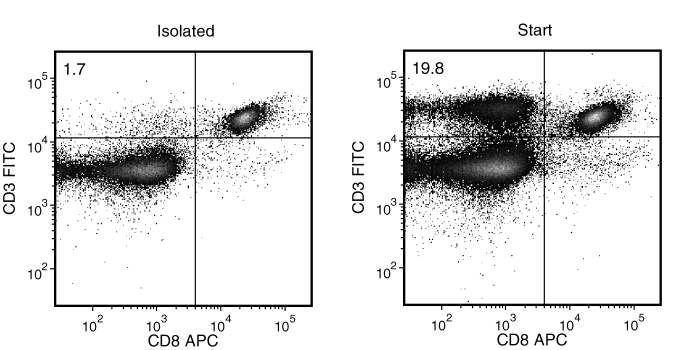
Figure 1. Typical Mouse Streptavidin Rapidspheres™ CD4 (CD3+CD8-) Depletion Profile
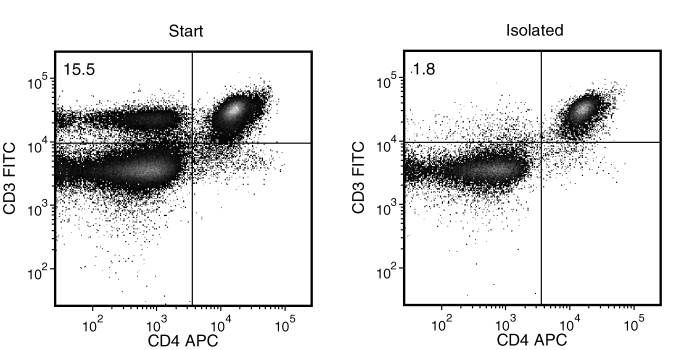
Figure 2. Typical Mouse Streptavidin Rapidspheres™ CD8 (CD3+CD4-) Depletion Profile
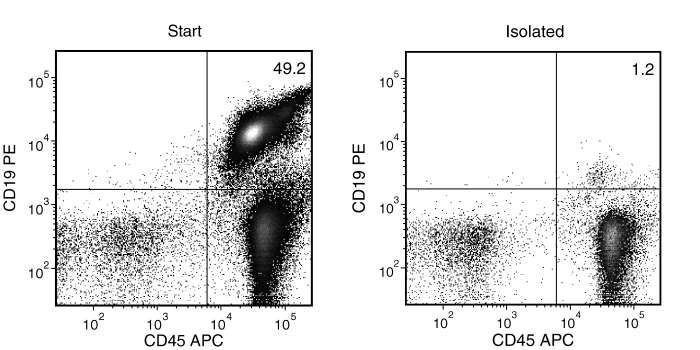
Figure 3. Typical Mouse Streptavidin Rapidspheres™ CD19 (CD19+CD45+) Depletion Profile
Publications (8)
Blood 2020 sep
Repurposing a novel anti-cancer RXR agonist to attenuate acute GVHD and maintain graft-versus-leukemia responses.
Abstract
Abstract
The nuclear receptors (NR) retinoid X receptors (RXRs) exert immunomodulatory functions to control inflammation and metabolism via homodimers and heterodimers with several other NRs including retinoic acid receptors. IRX4204 is a novel, highly specific RXR agonist in clinical trials that potently and selectively activates RXR homodimers but not heterodimers. Here, we show that in vivo IRX4204 was compared favorably with FK506 in abrogating acute graft-versus-host disease (GVHD), which was associated with inhibiting allogeneic donor T cell proliferation, reducing T helper 1 differentiation and promoting regulatory T cell (Treg) generation. Recipient IRX4204 treatment reduced intestinal injury and decreased IFN-$\gamma$ and TNF-$\alpha$ serum levels. Transcriptional analysis of donor T cells isolated from intestines of GVHD mice treated with IRX4204 revealed significant decreases in transcripts regulating pro-inflammatory pathways. In vitro, inducible Treg differentiation from na{\{i}}ve CD4+ T cells was enhanced by IRX4204; in vivo IRX4204 increased the conversion of donor Foxp3- T cells into peripheral Foxp3+ Tregs in GVHD mice. Using Foxp3 lineage tracer mice in which both the origin and current FoxP3 expression of Tregs can be tracked we demonstrate that IRX4204 supported Treg stability. Despite favoring Tregs and reducing Th1 differentiation IRX4204-treated recipients maintained graft-versus-leukemia responses against both leukemia and lymphoma cells. Notably IRX4204 reduced in vitro human T cell proliferation and enhanced Treg generation in mixed lymphocyte reaction cultures. Collectively these beneficial effects indicate that targeting RXRs with IRX4204 could be used as a novel approach to prevent acute GVHD in the clinic."
Molecular cancer research : MCR 2020 nov
Tumor-Resident Stromal Cells Promote Breast Cancer Invasion through Regulation of the Basal Phenotype.
Abstract
Abstract
Collective invasion can be led by breast cancer cells expressing basal epithelial markers, typified by keratin-14 (KRT14). We analyzed gene expression data from The Cancer Genome Atlas and demonstrated a significant correlation between a KRT14+ invasion signature and a stromal-mediated extracellular matrix (ECM) organization module. We then developed a novel coculture model of tumor organoids with autologous stromal cells. Coculture significantly increased KRT14 expression and invasion of organoids from both luminal and basal murine breast cancer models. However, stromal cell conditioned medium induced invasion but not KRT14 expression. Cancer cells released TGF$\beta$ and that signaling pathway was required for stromal cell-induced invasion and KRT14 expression. Mechanistically, TGF$\beta$ induced NOX4 expression in stromal cells and NOX4 inhibition reduced invasion and KRT14 expression. In summary, we developed a novel coculture model and revealed dynamic molecular interactions between stromal cells and cancer cells that regulate both basal gene expression and invasive behavior. IMPLICATIONS: Fibroblasts within mammary tumors can regulate the molecular phenotype and invasive behavior of breast cancer cells. VISUAL OVERVIEW: http://mcr.aacrjournals.org/content/molcanres/18/11/1615/F1.large.jpg.
Cell reports 2020 jun
Complete Topological Mapping of a Cellular Protein Interactome Reveals Bow-Tie Motifs as Ubiquitous Connectors of Protein Complexes.
Abstract
Abstract
The network topology of a protein interactome is shaped by the function of each protein, making it a resource of functional knowledge in tissues and in single cells. Today, this resource is underused, as complete network topology characterization has proved difficult for large protein interactomes. We apply a matrix visualization and decoding approach to a physical protein interactome of a dendritic cell, thereby characterizing its topology with no prior assumptions of structure. We discover 294 proteins, each forming topological motifs called bow-ties" that tie together the majority of observed protein complexes. The central proteins of these bow-ties have unique network properties display multifunctional capabilities are enriched for essential proteins and are widely expressed in other cells and tissues. Collectively the bow-tie motifs are a pervasive and previously unnoted topological trend in cellular interactomes. As such these results provide fundamental knowledge on how intracellular protein connectivity is organized and operates."
Nature medicine 2020 jul
Systemic dysfunction and plasticity of the immune macroenvironment in cancer models.
Abstract
Abstract
Understanding of the factors governing immune responses in cancer remains incomplete, limiting patient benefit. In this study, we used mass cytometry to define the systemic immune landscape in response to tumor development across five tissues in eight mouse tumor models. Systemic immunity was dramatically altered across models and time, with consistent findings in the peripheral blood of patients with breast cancer. Changes in peripheral tissues differed from those in the tumor microenvironment. Mice with tumor-experienced immune systems mounted dampened responses to orthogonal challenges, including reduced T cell activation during viral or bacterial infection. Antigen-presenting cells (APCs) mounted weaker responses in this context, whereas promoting APC activation rescued T cell activity. Systemic immune changes were reversed with surgical tumor resection, and many were prevented by interleukin-1 or granulocyte colony-stimulating factor blockade, revealing remarkable plasticity in the systemic immune state. These results demonstrate that tumor development dynamically reshapes the composition and function of the immune macroenvironment.
Cancer cell 2019 aug
Mechanisms of Progression of Myeloid Preleukemia to Transformed Myeloid Leukemia in Children with Down Syndrome.
Abstract
Abstract
Myeloid leukemia in Down syndrome (ML-DS) clonally evolves from transient abnormal myelopoiesis (TAM), a preleukemic condition in DS newborns. To define mechanisms of leukemic transformation, we combined exome and targeted resequencing of 111 TAM and 141 ML-DS samples with functional analyses. TAM requires trisomy 21 and truncating mutations in GATA1; additional TAM variants are usually not pathogenic. By contrast, in ML-DS, clonal and subclonal variants are functionally required. We identified a recurrent and oncogenic hotspot gain-of-function mutation in myeloid cytokine receptor CSF2RB. By a multiplex CRISPR/Cas9 screen in an in vivo murine TAM model, we tested loss-of-function of 22 recurrently mutated ML-DS genes. Loss of 18 different genes produced leukemias that phenotypically, genetically, and transcriptionally mirrored ML-DS.
Nature 2018 FEB
Population snapshots predict early haematopoietic and erythroid hierarchies.
Abstract
Abstract
The formation of red blood cells begins with the differentiation of multipotent haematopoietic progenitors. Reconstructing the steps of this differentiation represents a general challenge in stem-cell biology. Here we used single-cell transcriptomics, fate assays and a theory that allows the prediction of cell fates from population snapshots to demonstrate that mouse haematopoietic progenitors differentiate through a continuous, hierarchical structure into seven blood lineages. We uncovered coupling between the erythroid and the basophil or mast cell fates, a global haematopoietic response to erythroid stress and novel growth factor receptors that regulate erythropoiesis. We defined a flow cytometry sorting strategy to purify early stages of erythroid differentiation, completely isolating classically defined burst-forming and colony-forming progenitors. We also found that the cell cycle is progressively remodelled during erythroid development and during a sharp transcriptional switch that ends the colony-forming progenitor stage and activates terminal differentiation. Our work showcases the utility of linking transcriptomic data to predictive fate models, and provides insights into lineage development in vivo.