MethoCult™ H4435 Enriched
Methylcellulose-based medium with recombinant cytokines for human cells
概要
MethoCult™ H4435 Enriched (MethoCult™ GF+ H4435) is a complete methylcellulose-based medium for the growth and enumeration of hematopoietic progenitor cells in colony-forming unit (CFU) assays of human bone marrow, mobilized peripheral blood, peripheral blood, and cord blood samples. This medium supports optimal growth of erythroid progenitor cells (BFU-E and CFU-E), granulocyte-macrophage progenitor cells (CFU-GM, CFU-G and CFU-M), and multipotential granulocyte, erythroid, macrophage and megakaryocyte progenitor cells (CFU-GEMM). MethoCult™ H4435 Enriched is recommended for use with CD34+ cells and other purified cell populations, and for CFU assays performed as part of LTC-IC assays.
Browse our Frequently Asked Questions (FAQs) on performing the CFU assay and explore its utility as part of the cell therapy workflow.
Browse our Frequently Asked Questions (FAQs) on performing the CFU assay and explore its utility as part of the cell therapy workflow.
Contains
• Methylcellulose in Iscove's MDM
• Fetal bovine serum
• Bovine serum albumin
• 2-Mercaptoethanol
• Recombinant human stem cell factor (SCF)
• Recombinant human interleukin 3 (IL-3)
• Recombinant human interleukin 6 (IL-6)
• Recombinant human erythropoietin (EPO)
• Recombinant human granulocyte colony-stimulating factor (G-CSF)
• Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF)
• Supplements
• Fetal bovine serum
• Bovine serum albumin
• 2-Mercaptoethanol
• Recombinant human stem cell factor (SCF)
• Recombinant human interleukin 3 (IL-3)
• Recombinant human interleukin 6 (IL-6)
• Recombinant human erythropoietin (EPO)
• Recombinant human granulocyte colony-stimulating factor (G-CSF)
• Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF)
• Supplements
Subtype
Semi-Solid Media, Specialized Media
Cell Type
Hematopoietic Stem and Progenitor Cells
Species
Human, Non-Human Primate
Application
Cell Culture, Colony Assay, Functional Assay
Brand
MethoCult
Area of Interest
Stem Cell Biology
技术资料
| Document Type | 产品名称 | Catalog # | Lot # | 语言 |
|---|---|---|---|---|
| Product Information Sheet | MethoCult™ H4435 Enriched | 04435, 04445 | All | English |
| Manual | MethoCult™ H4435 Enriched | 04435 | All | English |
| Safety Data Sheet | MethoCult™ H4435 Enriched | 04435, 04445 | All | English |
数据及文献
Data
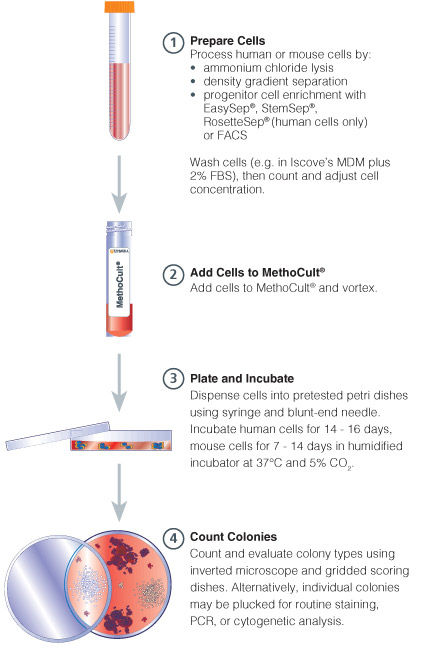
Figure 1. Procedure Summary for Hematopoietic CFU Assays
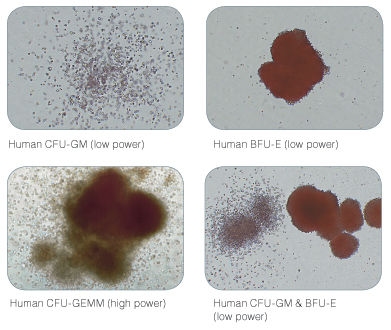
Figure 2. Examples of Colonies Derived from Human Hematopoietic Progenitors
Publications (34)
Advanced healthcare materials 2019 apr
A Human Hematopoietic Niche Model Supporting Hematopoietic Stem and Progenitor Cells In Vitro.
Abstract
Abstract
Niches in the bone marrow regulate hematopoietic stem and progenitor cell (HSPC) fate and behavior through cell-cell interactions and soluble factor secretion. The niche-HSPC crosstalk is a very complex process not completely elucidated yet. To aid further investigation of this crosstalk, a functional in vitro 3D model that closely represents the main supportive compartments of the bone marrow is developed. Different combinations of human stromal cells and hydrogels are tested for their potential to maintain CD34+ HSPCs. Cell viability, clonogenic hematopoietic potential, and surface marker expression are assessed over time. Optimal HSPC support is obtained in presence of adipogenic and osteogenic cells, together with progenitor derived endothelial cells. When cultured in a bioactive hydrogel, the supportive cells self-assemble into a hypoxic stromal network, stimulating CD34+ CD38+ cell formation, while maintaining the pool of CD34+ 38- HSPCs. HSPC clusters colocalize with the stromal networks, in close proximity to sinusoidal clusters of CD31+ endothelial cells. Importantly, the primary in vitro niche model supports HSPCs with no cytokine addition. Overall, the engineered primary 3D bone marrow environment provides an easy and reliable model to further investigate interactions between HSPCs and their endosteal and perivascular niches, in the context of normal hematopoiesis or blood-related diseases.
Nature protocols 2018 FEB
CRISPR/Cas9 genome editing in human hematopoietic stem cells.
Abstract
Abstract
Genome editing via homologous recombination (HR) (gene targeting) in human hematopoietic stem cells (HSCs) has the power to reveal gene-function relationships and potentially transform curative hematological gene and cell therapies. However, there are no comprehensive and reproducible protocols for targeting HSCs for HR. Herein, we provide a detailed protocol for the production, enrichment, and in vitro and in vivo analyses of HR-targeted HSCs by combining CRISPR/Cas9 technology with the use of rAAV6 and flow cytometry. Using this protocol, researchers can introduce single-nucleotide changes into the genome or longer gene cassettes with the precision of genome editing. Along with our troubleshooting and optimization guidelines, researchers can use this protocol to streamline HSC genome editing at any locus of interest. The in vitro HSC-targeting protocol and analyses can be completed in 3 weeks, and the long-term in vivo HSC engraftment analyses in immunodeficient mice can be achieved in 16 weeks. This protocol enables manipulation of genes for investigation of gene functions during hematopoiesis, as well as for the correction of genetic mutations in HSC transplantation-based therapies for diseases such as sickle cell disease, $\beta$-thalassemia, and primary immunodeficiencies.
Annals of hematology 2016 OCT
Generation of induced pluripotent stem cells as a potential source of hematopoietic stem cells for transplant in PNH patients.
Abstract
Abstract
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired hemolytic anemia caused by lack of CD55 and CD59 on blood cell membrane leading to increased sensitivity of blood cells to complement. Hematopoietic stem cell transplantation (HSCT) is the only curative therapy for PNH, however, lack of HLA-matched donors and post-transplant complications are major concerns. Induced pluripotent stem cells (iPSCs) derived from patients are an attractive source for generating autologous HSCs to avoid adverse effects resulting from allogeneic HSCT. The disease involves only HSCs and their progeny; therefore, other tissues are not affected by the mutation and may be used to produce disease-free autologous HSCs. This study aimed to derive PNH patient-specific iPSCs from human dermal fibroblasts (HDFs), characterize and differentiate to hematopoietic cells using a feeder-free protocol. Analysis of CD55 and CD59 expression was performed before and after reprogramming, and hematopoietic differentiation. Patients' dermal fibroblasts expressed CD55 and CD59 at normal levels and the normal expression remained after reprogramming. The iPSCs derived from PNH patients had typical pluripotent properties and differentiation capacities with normal karyotype. After hematopoietic differentiation, the differentiated cells expressed early hematopoietic markers (CD34 and CD43) with normal CD59 expression. The iPSCs derived from HDFs of PNH patients have normal levels of CD55 and CD59 expression and hold promise as a potential source of HSCs for autologous transplantation to cure PNH patients.
Cell and Tissue Research 2016 JUL
Cell type of origin influences iPSC generation and differentiation to cells of the hematoendothelial lineage
Abstract
Abstract
The use of induced pluripotent stem cells (iPSCs) as a source of cells for cell-based therapy in regenerative medicine is hampered by the limited efficiency and safety of the reprogramming procedure and the low efficiency of iPSC differentiation to specialized cell types. Evidence suggests that iPSCs retain an epigenetic memory of their parental cells with a possible influence on their differentiation capacity in vitro. We reprogramme three cell types, namely human umbilical cord vein endothelial cells (HUVECs), endothelial progenitor cells (EPCs) and human dermal fibroblasts (HDFs), to iPSCs and compare their hematoendothelial differentiation capacity. HUVECs and EPCs were at least two-fold more efficient in iPSC reprogramming than HDFs. Both HUVEC- and EPC-derived iPSCs exhibited high potentiality toward endothelial cell differentiation compared with HDF-derived iPSCs. However, only HUVEC-derived iPSCs showed efficient differentiation to hematopoietic stem/progenitor cells. Examination of DNA methylation at promoters of hematopoietic and endothelial genes revealed evidence for the existence of epigenetic memory at the endothelial genes but not the hematopoietic genes in iPSCs derived from HUVECs and EPCs indicating that epigenetic memory involves an endothelial differentiation bias. Our findings suggest that endothelial cells and EPCs are better sources for iPSC derivation regarding their reprogramming efficiency and that the somatic cell type used for iPSC generation toward specific cell lineage differentiation is of importance.
Nature Biotechnology 2016 APR
Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity.
Abstract
Abstract
Current combination antiretroviral therapies (cART) efficiently suppress HIV-1 reproduction in humans, but the virus persists as integrated proviral reservoirs in small numbers of cells. To generate an antiviral agent capable of eradicating the provirus from infected cells, we employed 145 cycles of substrate-linked directed evolution to evolve a recombinase (Brec1) that site-specifically recognizes a 34-bp sequence present in the long terminal repeats (LTRs) of the majority of the clinically relevant HIV-1 strains and subtypes. Brec1 efficiently, precisely and safely removes the integrated provirus from infected cells and is efficacious on clinical HIV-1 isolates in vitro and in vivo, including in mice humanized with patient-derived cells. Our data suggest that Brec1 has potential for clinical application as a curative HIV-1 therapy.
Journal of Biological Chemistry 2015 MAY
Factor-induced Reprogramming and Zinc Finger Nuclease-aided Gene Targeting Cause Different Genome Instability in $\$-Thalassemia Induced Pluripotent Stem Cells (iPSCs).
Abstract
Abstract
The generation of personalized induced pluripotent stem cells (iPSCs) followed by targeted genome editing provides an opportunity for developing customized effective cellular therapies for genetic disorders. However, it is critical to ascertain whether edited iPSCs harbor unfavorable genomic variations before their clinical application. To examine the mutation status of the edited iPSC genome and trace the origin of possible mutations at different steps, we have generated virus-free iPSCs from amniotic cells carrying homozygous point mutations in beta-hemoglobin gene (HBB) that cause severe beta-thalassemia (beta-Thal), corrected the mutations in both HBB alleles by zinc finger nuclease-aided gene targeting, and obtained the final HBB gene-corrected iPSCs by excising the exogenous drug resistance gene with Cre recombinase. Through comparative genomic hybridization and whole-exome sequencing, we uncovered seven copy number variations, five small insertions/deletions, and 64 single nucleotide variations (SNVs) in beta-Thal iPSCs before the gene targeting step and found a single small copy number variation, 19 insertions/deletions, and 340 single nucleotide variations in the final gene-corrected beta-Thal iPSCs. Our data revealed that substantial but different genomic variations occurred at factor-induced somatic cell reprogramming and zinc finger nuclease-aided gene targeting steps, suggesting that stringent genomic monitoring and selection are needed both at the time of iPSC derivation and after gene targeting.