ClonaCell™-HY Hybridoma Kit
Complete kit for hybridoma generation
概要
ClonaCell™-HY Hybridoma Kit contains media and reagents for all steps in hybridoma development and monoclonal antibody production. The ClonaCell™-HY method uses a methylcellulose-based semi-solid selective medium to combine hybridoma selection and cloning into one step. Individual parental hybridoma clones and their progeny remain localized together in the semi-solid medium as they grow to form distinct colonies. This prevents the loss of rare clones by overgrowth from faster-growing cells, as can occur during selection in a liquid medium. The hybridoma colonies can be easily picked from the semi-solid medium by manual or robotic methods and dispersed into a liquid growth medium for screening and expansion. This kit has been verified for use in mouse and rat hydridoma development and monoclonal antibody production and reportedly is compatible for production, cloning, and expansion of hybridomas using lymphocytes from a variety of host animals including human, mouse, rat, and hamster.
Advantages
• Time savings: Hybridoma selection and cloning are combined into one step
• Resource savings: Single cell-derived hybridomas form visible discrete colonies in semi-solid medium, and are easy to pick and screen, and transfer to liquid medium for expansion
• Cloning efficiency: Individual cells are suspended and immobilized in semi-solid medium, preventing loss of rare, high-producing clones by overgrowth
• Resource savings: Single cell-derived hybridomas form visible discrete colonies in semi-solid medium, and are easy to pick and screen, and transfer to liquid medium for expansion
• Cloning efficiency: Individual cells are suspended and immobilized in semi-solid medium, preventing loss of rare, high-producing clones by overgrowth
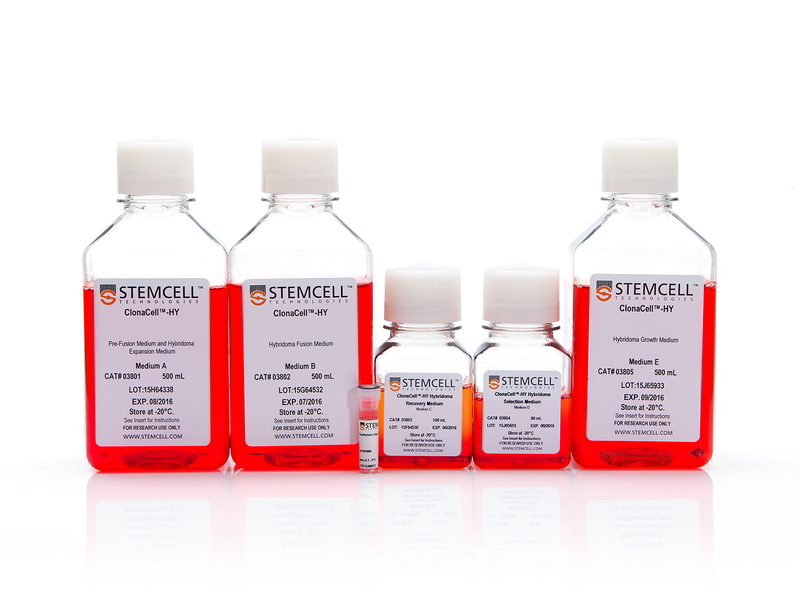
Components
- ClonaCell™-HY Medium A, 500 mL (Catalog #03801)
- ClonaCell™-HY Medium B, 500 mL (Catalog #03802)
- ClonaCell™-HY Medium C, 100 mL (Catalog #03803)
- ClonaCell™-HY Medium D, 90 mL (Catalog #03804)
- ClonaCell™-HY Medium E, 500 mL (Catalog #03805)
- ClonaCell™-HY PEG, 1.5 mL (Catalog #03806)
Subtype
Semi-Solid Media, Specialized Media
Cell Type
Hybridomas
Species
Mouse, Rat, Other
Application
Cell Culture, Hybridoma Generation
Brand
ClonaCell
Area of Interest
Antibody Development, Cell Line Development, Drug Discovery and Toxicity Testing, Hybridoma Generation
技术资料
数据及文献
Publications (67)
Proceedings of the National Academy of Sciences of the United States of America 2016 OCT
Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein.
Abstract
Abstract
Palivizumab was the first antiviral monoclonal antibody (mAb) approved for therapeutic use in humans, and remains a prophylactic treatment for infants at risk for severe disease because of respiratory syncytial virus (RSV). Palivizumab is an engineered humanized version of a murine mAb targeting antigenic site II of the RSV fusion (F) protein, a key target in vaccine development. There are limited reported naturally occurring human mAbs to site II; therefore, the structural basis for human antibody recognition of this major antigenic site is poorly understood. Here, we describe a nonneutralizing class of site II-specific mAbs that competed for binding with palivizumab to postfusion RSV F protein. We also describe two classes of site II-specific neutralizing mAbs, one of which escaped competition with nonneutralizing mAbs. An X-ray crystal structure of the neutralizing mAb 14N4 in complex with F protein showed that the binding angle at which human neutralizing mAbs interact with antigenic site II determines whether or not nonneutralizing antibodies compete with their binding. Fine-mapping studies determined that nonneutralizing mAbs that interfere with binding of neutralizing mAbs recognize site II with a pose that facilitates binding to an epitope containing F surface residues on a neighboring protomer. Neutralizing antibodies, like motavizumab and a new mAb designated 3J20 that escape interference by the inhibiting mAbs, avoid such contact by binding at an angle that is shifted away from the nonneutralizing site. Furthermore, binding to rationally and computationally designed site II helix-loop-helix epitope-scaffold vaccines distinguished neutralizing from nonneutralizing site II antibodies.
Nature 2016 NOV
Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice.
Abstract
Abstract
Zika virus (ZIKV) is an emerging mosquito-transmitted flavivirus that can cause severe disease, including congenital birth defects during pregnancy(1). To develop candidate therapeutic agents against ZIKV, we isolated a panel of human monoclonal antibodies (mAbs) from subjects with prior ZIKV infection. A subset of mAbs recognized diverse epitopes on the envelope (E) protein and exhibited potently neutralizing activity. One of the most inhibitory mAbs, ZIKV-117, broadly neutralized infection of ZIKV strains corresponding to African, Asian, and American lineages. Epitope mapping studies revealed that ZIKV-117 recognized a unique quaternary epitope on the E protein dimer-dimer interface. We evaluated the therapeutic efficacy of ZIKV-117 in pregnant and non-pregnant mice. mAb treatment markedly reduced tissue pathology, placental and fetal infection, and mortality in mice. Thus, neutralizing human mAbs can protect against maternal-fetal transmission, infection and disease, and reveal important determinants for structure-based rational vaccine design efforts.
Antiviral Research 2016 NOV
An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo
Abstract
Abstract
Zoonotic A(H7N9) avian influenza viruses emerged in China in 2013 and continue to be a threat to human public health, having infected over 800 individuals with a mortality rate approaching 40%. Treatment options for people infected with A(H7N9) include the use of neuraminidase (NA) inhibitors. However, like other influenza viruses, A(H7N9) can become resistant to these drugs. The use of monoclonal antibodies is a rapidly developing strategy for controlling influenza virus infection. Here we generated a murine monoclonal antibody (3c10-3) directed against the NA of A(H7N9) and show that prophylactic systemic administration of 3c10-3 fully protected mice from lethal challenge with wild-type A/Anhui/1/2013 (H7N9). Further, post-infection treatment with a single systemic dose of 3c10-3 at either 24, 48 or 72 h post A(H7N9) challenge resulted in both dose- and time-dependent protection of up to 100% of mice, demonstrating therapeutic potential for 3c10-3. Epitope mapping revealed that 3c10-3 binds near the enzyme active site of NA, and functional characterization showed that 3c10-3 inhibits the enzyme activity of NA and restricts the cell-to-cell spread of the virus in cultured cells. Affinity analysis also revealed that 3c10-3 binds equally well to recombinant NA of wild-type A/Anhui/1/2013 and to a variant NA carrying a R289K mutation known to infer NAI resistance. These results suggest that 3c10-3 has the potential to be used as a therapeutic to treat A(H7N9) infections either as an alternative to, or in combination with, current NA antiviral inhibitors.
The Journal of Immunology 2016 MAY
Immunodominant West Nile virus T cell epitopes are fewer in number and fashionably late
Abstract
Abstract
Class I HLA molecules mark infected cells for immune targeting by presenting pathogen-encoded peptides on the cell surface. Characterization of viral peptides unique to infected cells is important for understanding CD8(+) T cell responses and for the development of T cell-based immunotherapies. Having previously reported a series of West Nile virus (WNV) epitopes that are naturally presented by HLA-A*02:01, in this study we generated TCR mimic (TCRm) mAbs to three of these peptide/HLA complexes-the immunodominant SVG9 (E protein), the subdominant SLF9 (NS4B protein), and the immunorecessive YTM9 (NS3 protein)-and used these TCRm mAbs to stain WNV-infected cell lines and primary APCs. TCRm staining of WNV-infected cells demonstrated that the immunorecessive YTM9 appeared several hours earlier and at 5- to 10-fold greater density than the more immunogenic SLF9 and SVG9 ligands, respectively. Moreover, staining following inhibition of the TAP demonstrated that all three viral ligands were presented in a TAP-dependent manner despite originating from different cellular compartments. To our knowledge, this study represents the first use of TCRm mAbs to define the kinetics and magnitude of HLA presentation for a series of epitopes encoded by one virus, and the results depict a pattern whereby individual epitopes differ considerably in abundance and availability. The observations that immunodominant ligands can be found at lower levels and at later time points after infection suggest that a reevaluation of the factors that combine to shape T cell reactivity may be warranted.
mBio 2016 MAY
Deletion of a Yci1 Domain Protein of Candida albicans Allows Homothallic Mating in MTL Heterozygous Cells
Abstract
Abstract
It has been proposed that the ancestral fungus was mating competent and homothallic. However, many mating-competent fungi were initially classified as asexual because their mating capacity was hidden behind layers of regulation. For efficient in vitro mating, the essentially obligate diploid ascomycete pathogen Candida albicans has to change its mating type locus from heterozygous MTL a /α to homozygous MTL a / a or MTL α/α and then undergo an environmentally controlled epigenetic switch to the mating-competent opaque form. These requirements greatly reduce the potential for C. albicans mating. Deletion of the Yci1 domain gene OFR1 bypasses the need for C. albicans cells to change the mating type locus from heterozygous to homozygous prior to switching to the opaque form and mating and allows homothallic mating of MTL heterozygous strains. This bypass is carbon source dependent and does not occur when cells are grown on glucose. Transcriptional profiling of ofr1 mutant cells shows that in addition to regulating cell type and mating circuitry, Ofr1 is needed for proper regulation of histone and chitin biosynthesis gene expression. It appears that OFR1 is a key regulator in C. albicans and functions in part to maintain the cryptic mating phenotype of the pathogen.
Human Molecular Genetics 2016 MAY
Characterization of SKAP/kinastrin isoforms: the N-terminus defines tissue specificity and Pontin binding
Abstract
Abstract
Small Kinetochore-Associated Protein (SKAP)/Kinastrin is a multifunctional protein with proposed roles in mitosis, apoptosis and cell migration. Exact mechanisms underlying its activities in these cellular processes are not completely understood. SKAP is predicted to have different isoforms, however, previous studies did not differentiate between them. Since distinct molecular architectures of protein isoforms often influence their localization and functions, this study aimed to examine the expression profile and functional differences between SKAP isoforms in human and mouse. Analyses of various human tissues and cells of different origin by RT-PCR, and by Western blotting and immunocytochemistry applying newly generated anti-SKAP monoclonal antibodies revealed that human SKAP exists in two protein isoforms: ubiquitously expressed SKAP16 and testis/sperm-specific SKAP1. In mouse, SKAP1 expression is detectable in testis at 4 weeks postnatally, when the first wave of spermatogenesis in mice is complete and the elongated spermatids are present in the testes. Furthermore, we identified Pontin as a new SKAP1 interaction partner. SKAP1 and Pontin co-localized in the flagellar region of human sperm suggesting a functional relevance for SKAP1-Pontin interaction in sperm motility. Since most previous studies on SKAP were performed with the testis-specific isoform SKAP1, our findings provide a new basis for future studies on the role of SKAP in both human somatic cells and male germ cells, including studies on male fertility.